import geopandas as gpd
import foliumSpatial Visualization with Python
Loading packages
Load and inspect meuse data
meuse = gpd.read_file("meuse.shp") # Adjust the file path as needed
# Inspect the first few rows of the data
print(meuse.head()) cadmium copper lead zinc elev dist om ffreq soil lime \
0 11.7 85.0 299.0 1022.0 7.909 0.001358 13.6 1 1 1
1 8.6 81.0 277.0 1141.0 6.983 0.012224 14.0 1 1 1
2 6.5 68.0 199.0 640.0 7.800 0.103029 13.0 1 1 1
3 2.6 81.0 116.0 257.0 7.655 0.190094 8.0 1 2 0
4 2.8 48.0 117.0 269.0 7.480 0.277090 8.7 1 2 0
landuse dist_m geometry
0 Ah 50.0 POINT (181072.000 333611.000)
1 Ah 30.0 POINT (181025.000 333558.000)
2 Ah 150.0 POINT (181165.000 333537.000)
3 Ga 270.0 POINT (181298.000 333484.000)
4 Ah 380.0 POINT (181307.000 333330.000) Visualise the data
import matplotlib.pyplot as plt
# Plot the meuse dataset (points in this case)
meuse.plot(marker='o', color='blue', markersize=5)
# Show the plot
plt.title('Meuse Spatial Data Visualization')
plt.xlabel('Longitude')
plt.ylabel('Latitude')
plt.show()
Customize plot
meuse.plot(column='zinc', cmap='viridis', legend=True, markersize=5)
# Show the plot
plt.title('Zinc Concentration in Meuse Data')
plt.xlabel('Longitude')
plt.ylabel('Latitude')
plt.show()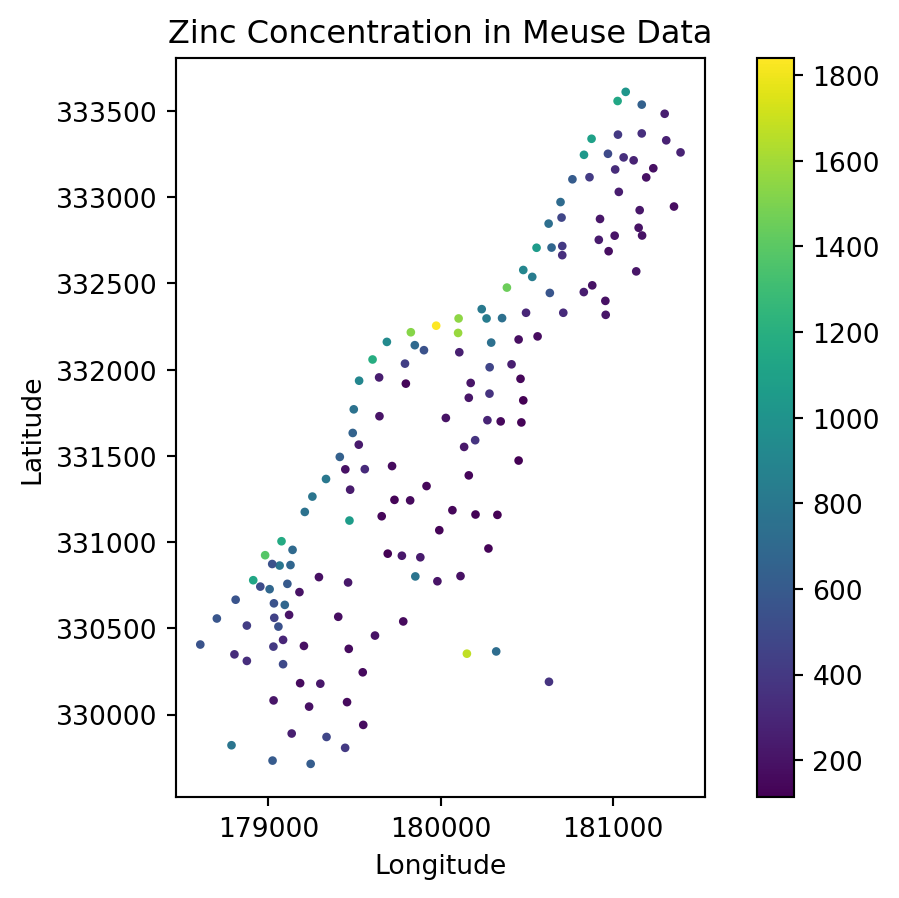
# Ensure the GeoDataFrame has the correct CRS and reproject to WGS84 for folium
gdf = meuse.to_crs(epsg=4326)
# Create a folium map centered on the data
map_center = [gdf.geometry.y.mean(), gdf.geometry.x.mean()]
m = folium.Map(location=map_center, zoom_start=12)
# Add points to the map with a popup for one of the attributes
for _, row in gdf.iterrows():
folium.CircleMarker(
location=[row.geometry.y, row.geometry.x],
radius=5,
color='blue',
fill=True,
fill_opacity=0.6,
popup=f"Zinc: {row['zinc']}" # Replace 'zinc' with a column from your shapefile
).add_to(m)
# Save or display the map
m.save("meuse_map.html")
mMake this Notebook Trusted to load map: File -> Trust Notebook