library(tidymodels)
library(tidyverse)
library(gt)
library(stringr)
library(visdat)
library(GGally)8 Predictive modelling: Measures of Accuracy
8.1 Packages
8.2 Import Data and Check Data Format
link <- "https://raw.githubusercontent.com/kirenz/datasets/master/housing_unclean.csv"
hdf <- read_csv(link) #readrRows: 20640 Columns: 10
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (3): housing_median_age, median_house_value, ocean_proximity
dbl (7): longitude, latitude, total_rooms, total_bedrooms, population, house...
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.dim(hdf)[1] 20640 10colnames(hdf) [1] "longitude" "latitude" "housing_median_age"
[4] "total_rooms" "total_bedrooms" "population"
[7] "households" "median_income" "median_house_value"
[10] "ocean_proximity" #View(hdf)
head(hdf) |>
gt()| longitude | latitude | housing_median_age | total_rooms | total_bedrooms | population | households | median_income | median_house_value | ocean_proximity |
|---|---|---|---|---|---|---|---|---|---|
| -122.23 | 37.88 | 41.0years | 880 | 129 | 322 | 126 | 8.3252 | 452600.0$ | NEAR BAY |
| -122.22 | 37.86 | 21.0 | 7099 | 1106 | 2401 | 1138 | 8.3014 | 358500.0 | NEAR BAY |
| -122.24 | 37.85 | 52.0 | 1467 | 190 | 496 | 177 | 7.2574 | 352100.0 | NEAR BAY |
| -122.25 | 37.85 | 52.0 | 1274 | 235 | 558 | 219 | 5.6431 | 341300.0 | NEAR BAY |
| -122.25 | 37.85 | 52.0 | 1627 | 280 | 565 | 259 | 3.8462 | 342200.0 | NEAR BAY |
| -122.25 | 37.85 | 52.0 | 919 | 213 | 413 | 193 | 4.0368 | 269700.0 | NEAR BAY |
hdf |>
slice_head(n=6) |>
gt()| longitude | latitude | housing_median_age | total_rooms | total_bedrooms | population | households | median_income | median_house_value | ocean_proximity |
|---|---|---|---|---|---|---|---|---|---|
| -122.23 | 37.88 | 41.0years | 880 | 129 | 322 | 126 | 8.3252 | 452600.0$ | NEAR BAY |
| -122.22 | 37.86 | 21.0 | 7099 | 1106 | 2401 | 1138 | 8.3014 | 358500.0 | NEAR BAY |
| -122.24 | 37.85 | 52.0 | 1467 | 190 | 496 | 177 | 7.2574 | 352100.0 | NEAR BAY |
| -122.25 | 37.85 | 52.0 | 1274 | 235 | 558 | 219 | 5.6431 | 341300.0 | NEAR BAY |
| -122.25 | 37.85 | 52.0 | 1627 | 280 | 565 | 259 | 3.8462 | 342200.0 | NEAR BAY |
| -122.25 | 37.85 | 52.0 | 919 | 213 | 413 | 193 | 4.0368 | 269700.0 | NEAR BAY |
8.3 Preprocessing
hdf <-
hdf |>
mutate(
housing_median_age=str_remove_all(housing_median_age, "[years]"),
median_house_value = str_remove_all(median_house_value, "[$]"))
#View(hdf)8.3.1 Recheck the format
glimpse(hdf) Rows: 20,640
Columns: 10
$ longitude <dbl> -122.23, -122.22, -122.24, -122.25, -122.25, -122.2…
$ latitude <dbl> 37.88, 37.86, 37.85, 37.85, 37.85, 37.85, 37.84, 37…
$ housing_median_age <chr> "41.0", "21.0", "52.0", "52.0", "52.0", "52.0", "52…
$ total_rooms <dbl> 880, 7099, 1467, 1274, 1627, 919, 2535, 3104, 2555,…
$ total_bedrooms <dbl> 129, 1106, 190, 235, 280, 213, 489, 687, 665, 707, …
$ population <dbl> 322, 2401, 496, 558, 565, 413, 1094, 1157, 1206, 15…
$ households <dbl> 126, 1138, 177, 219, 259, 193, 514, 647, 595, 714, …
$ median_income <dbl> 8.3252, 8.3014, 7.2574, 5.6431, 3.8462, 4.0368, 3.6…
$ median_house_value <chr> "452600.0", "358500.0", "352100.0", "341300.0", "34…
$ ocean_proximity <chr> "NEAR BAY", "NEAR BAY", "NEAR BAY", "NEAR BAY", "NE…8.3.2 Visualize the structure
vis_dat(hdf)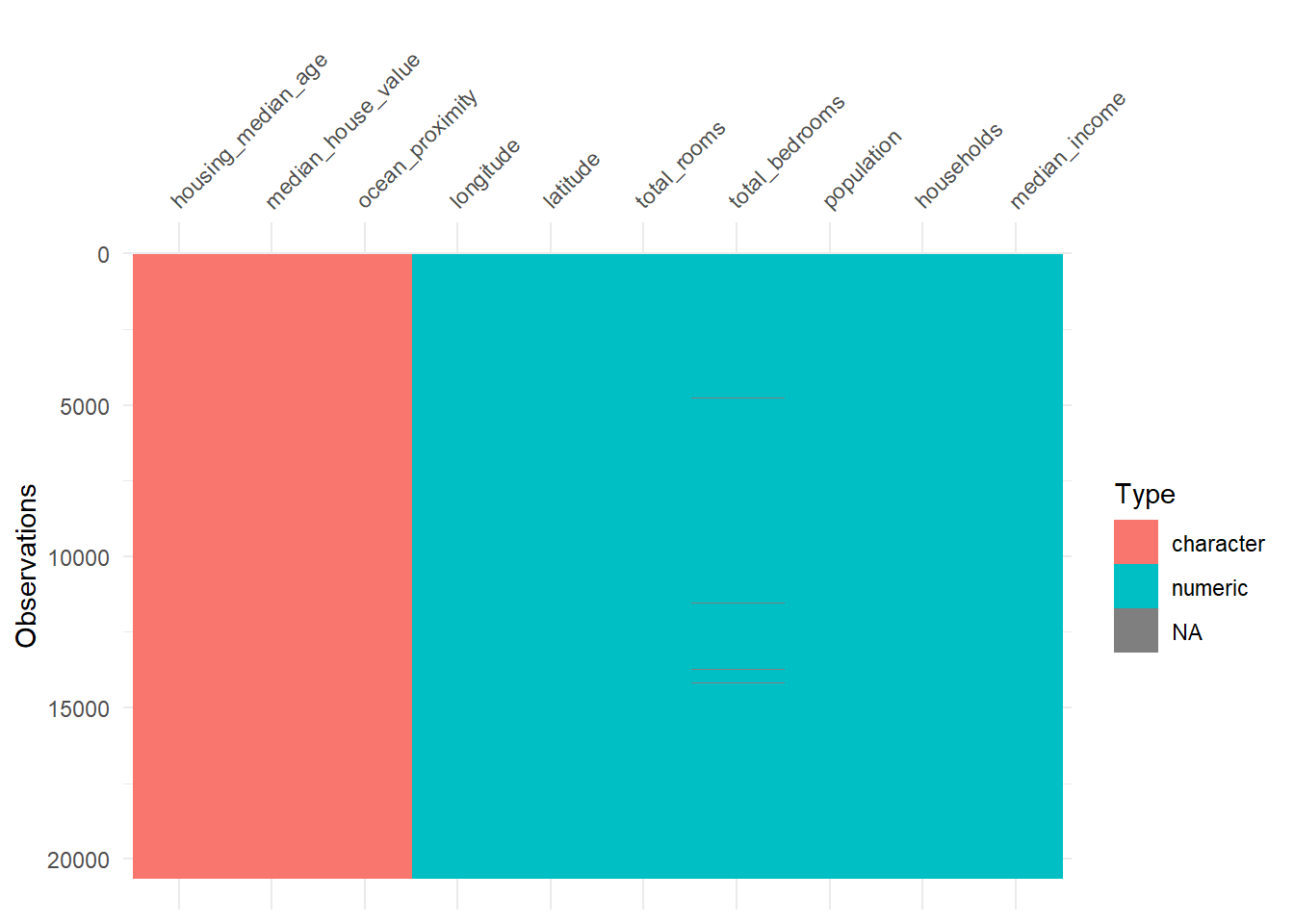
8.3.3 Observe summary statistics
hdf |>
count(ocean_proximity, sort=TRUE)# A tibble: 5 × 2
ocean_proximity n
<chr> <int>
1 <1H OCEAN 9136
2 INLAND 6551
3 NEAR OCEAN 2658
4 NEAR BAY 2290
5 ISLAND 58.3.4 Convert qualitative variables into factor
hdf <- hdf |>
mutate(across(where(is.character), as.factor))
vis_dat(hdf)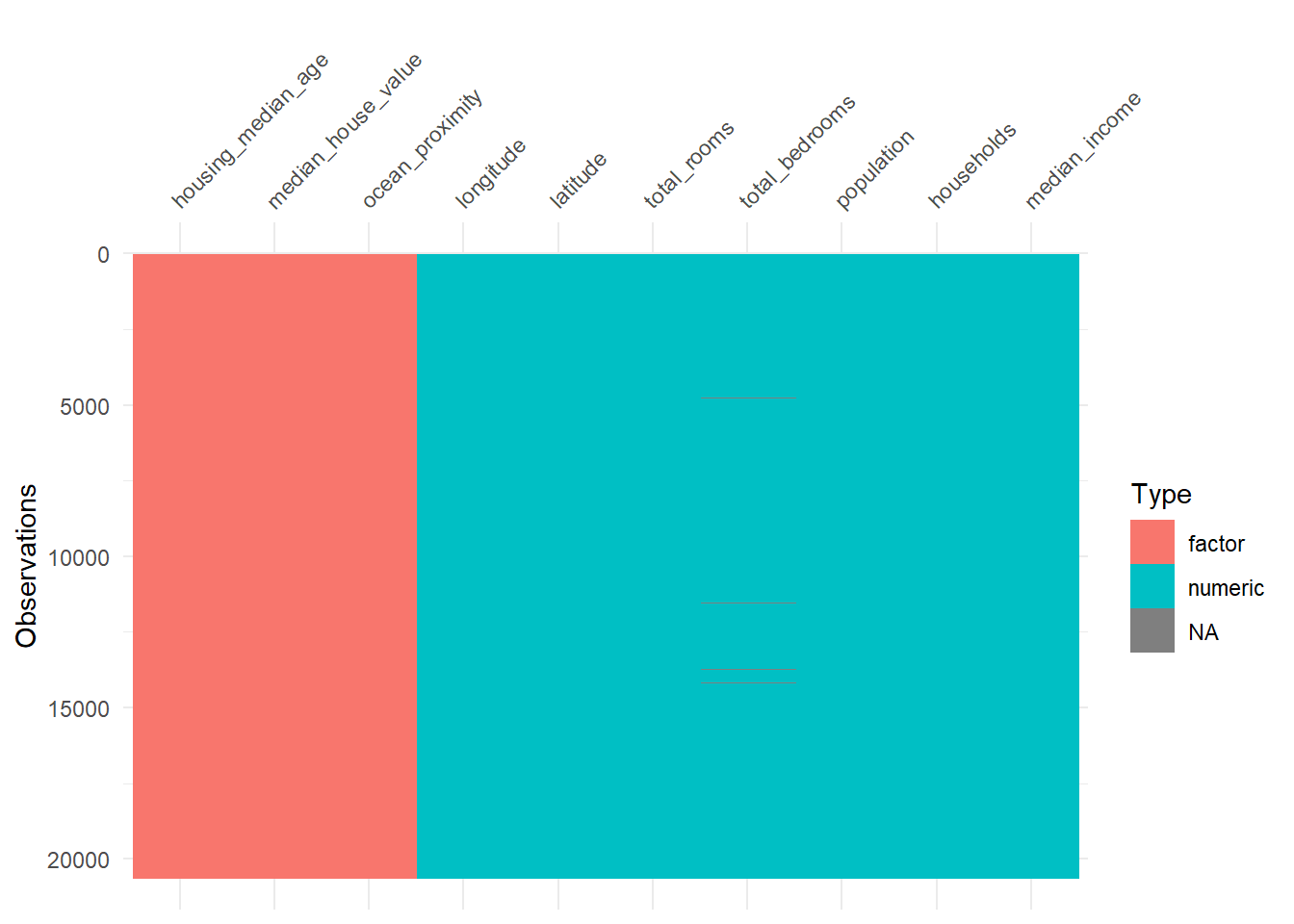
8.3.5 Convert variables into numeric
hdf <- hdf |>
mutate(
housing_median_age = as.numeric(housing_median_age),
median_house_value = as.numeric(median_house_value)
)
vis_dat(hdf)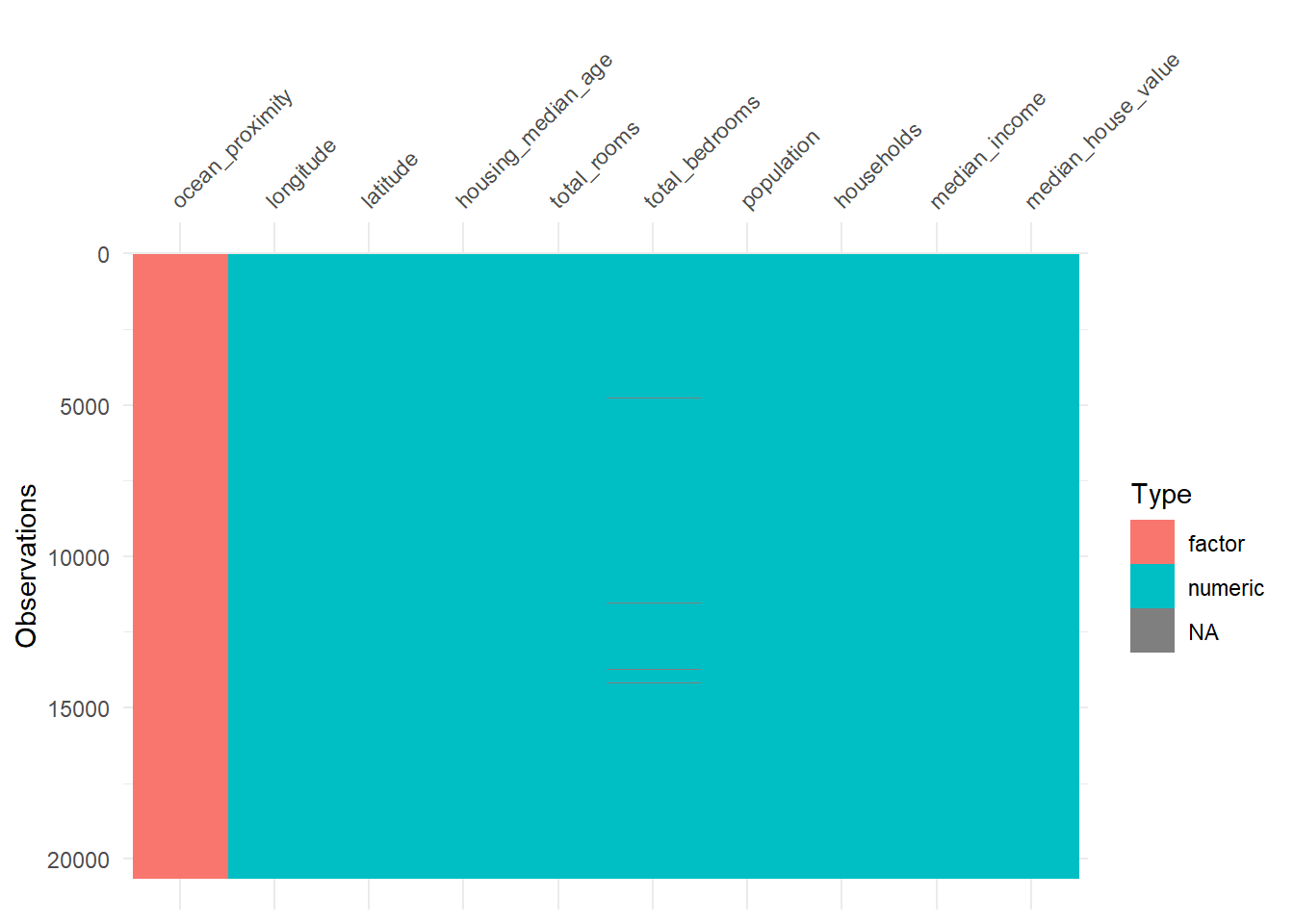
8.3.6 Observe missing value
vis_miss(hdf, sort_miss=TRUE)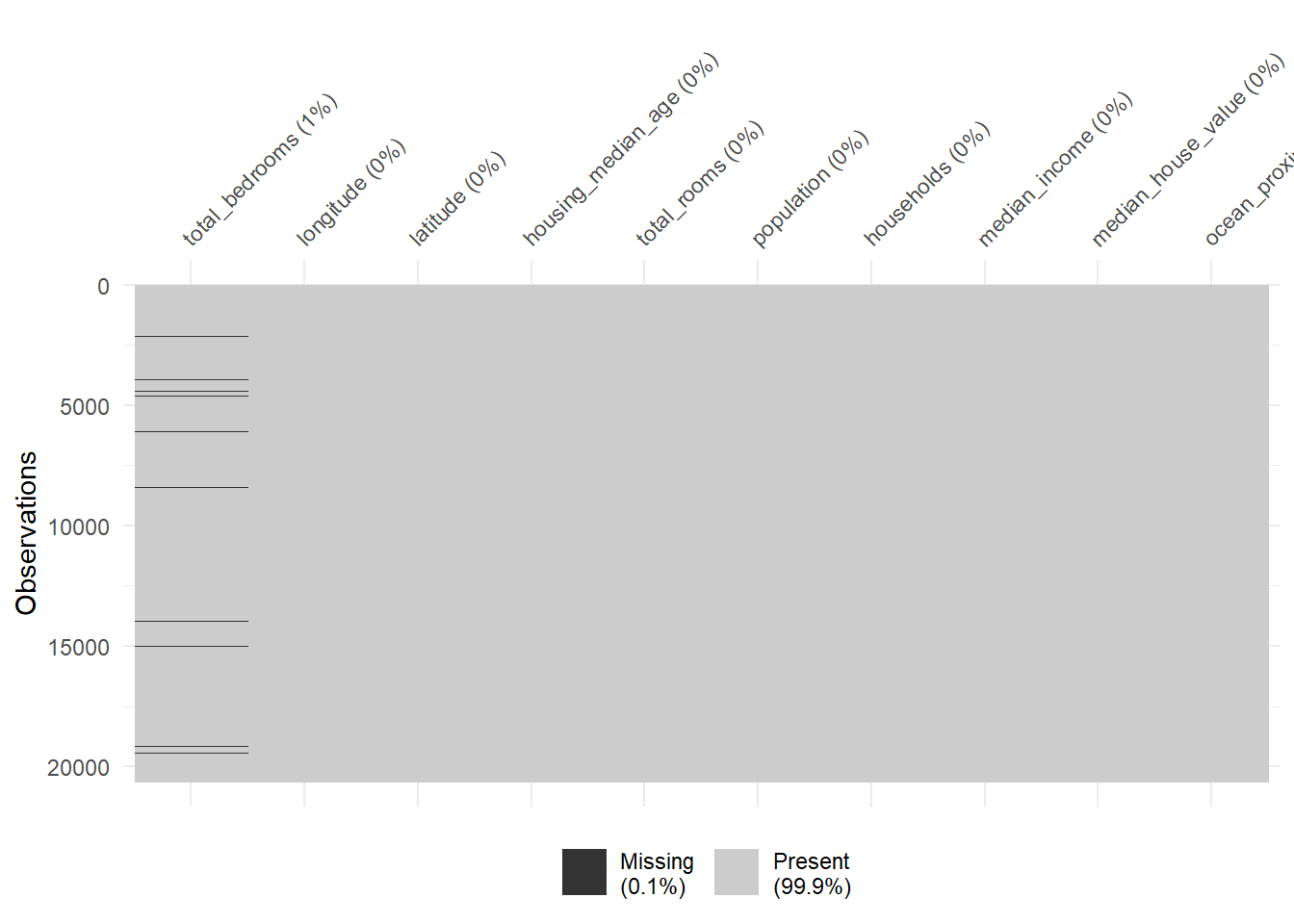
is.na(hdf) |> colSums() longitude latitude housing_median_age total_rooms
0 0 0 0
total_bedrooms population households median_income
207 0 0 0
median_house_value ocean_proximity
0 0 8.4 Create new variables
hdf <- hdf |>
mutate(rooms_per_household = total_rooms/households,
bedrooms_per_room = total_bedrooms/total_rooms,
population_per_household = population/households)
#View(hdf)8.5 Categorize the dependent variable
hdf <- hdf |>
mutate(price_category = case_when(
median_house_value < 1500 ~ "below",
median_house_value >= 1500 ~ "above",
)) |>
mutate(price_category = as.factor(price_category)) |>
select(-median_house_value)
#View(hdf)8.6 Distribution of the dependent variable
hdf |>
count(price_category, name="count") |>
mutate(percent = count/sum(count)) |>
gt()| price_category | count | percent |
|---|---|---|
| above | 9364 | 0.4536822 |
| below | 11276 | 0.5463178 |
8.7 Check the structure again
skimr::skim(hdf)| Name | hdf |
| Number of rows | 20640 |
| Number of columns | 13 |
| _______________________ | |
| Column type frequency: | |
| factor | 2 |
| numeric | 11 |
| ________________________ | |
| Group variables | None |
Variable type: factor
| skim_variable | n_missing | complete_rate | ordered | n_unique | top_counts |
|---|---|---|---|---|---|
| ocean_proximity | 0 | 1 | FALSE | 5 | <1H: 9136, INL: 6551, NEA: 2658, NEA: 2290 |
| price_category | 0 | 1 | FALSE | 2 | bel: 11276, abo: 9364 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| longitude | 0 | 1.00 | -119.57 | 2.00 | -124.35 | -121.80 | -118.49 | -118.01 | -114.31 | ▂▆▃▇▁ |
| latitude | 0 | 1.00 | 35.63 | 2.14 | 32.54 | 33.93 | 34.26 | 37.71 | 41.95 | ▇▁▅▂▁ |
| housing_median_age | 0 | 1.00 | 25.01 | 13.48 | 1.00 | 14.00 | 25.00 | 35.00 | 52.00 | ▆▆▇▅▅ |
| total_rooms | 0 | 1.00 | 2635.76 | 2181.62 | 2.00 | 1447.75 | 2127.00 | 3148.00 | 39320.00 | ▇▁▁▁▁ |
| total_bedrooms | 207 | 0.99 | 537.87 | 421.39 | 1.00 | 296.00 | 435.00 | 647.00 | 6445.00 | ▇▁▁▁▁ |
| population | 0 | 1.00 | 1425.48 | 1132.46 | 3.00 | 787.00 | 1166.00 | 1725.00 | 35682.00 | ▇▁▁▁▁ |
| households | 0 | 1.00 | 499.54 | 382.33 | 1.00 | 280.00 | 409.00 | 605.00 | 6082.00 | ▇▁▁▁▁ |
| median_income | 0 | 1.00 | 3.87 | 1.90 | 0.50 | 2.56 | 3.53 | 4.74 | 15.00 | ▇▇▁▁▁ |
| rooms_per_household | 0 | 1.00 | 5.43 | 2.47 | 0.85 | 4.44 | 5.23 | 6.05 | 141.91 | ▇▁▁▁▁ |
| bedrooms_per_room | 207 | 0.99 | 0.21 | 0.06 | 0.10 | 0.18 | 0.20 | 0.24 | 1.00 | ▇▁▁▁▁ |
| population_per_household | 0 | 1.00 | 3.07 | 10.39 | 0.69 | 2.43 | 2.82 | 3.28 | 1243.33 | ▇▁▁▁▁ |
8.8 EDA
hdf |>
select(housing_median_age,
median_income,
bedrooms_per_room,
rooms_per_household,
population_per_household) |>
ggscatmat(alpha=0.2)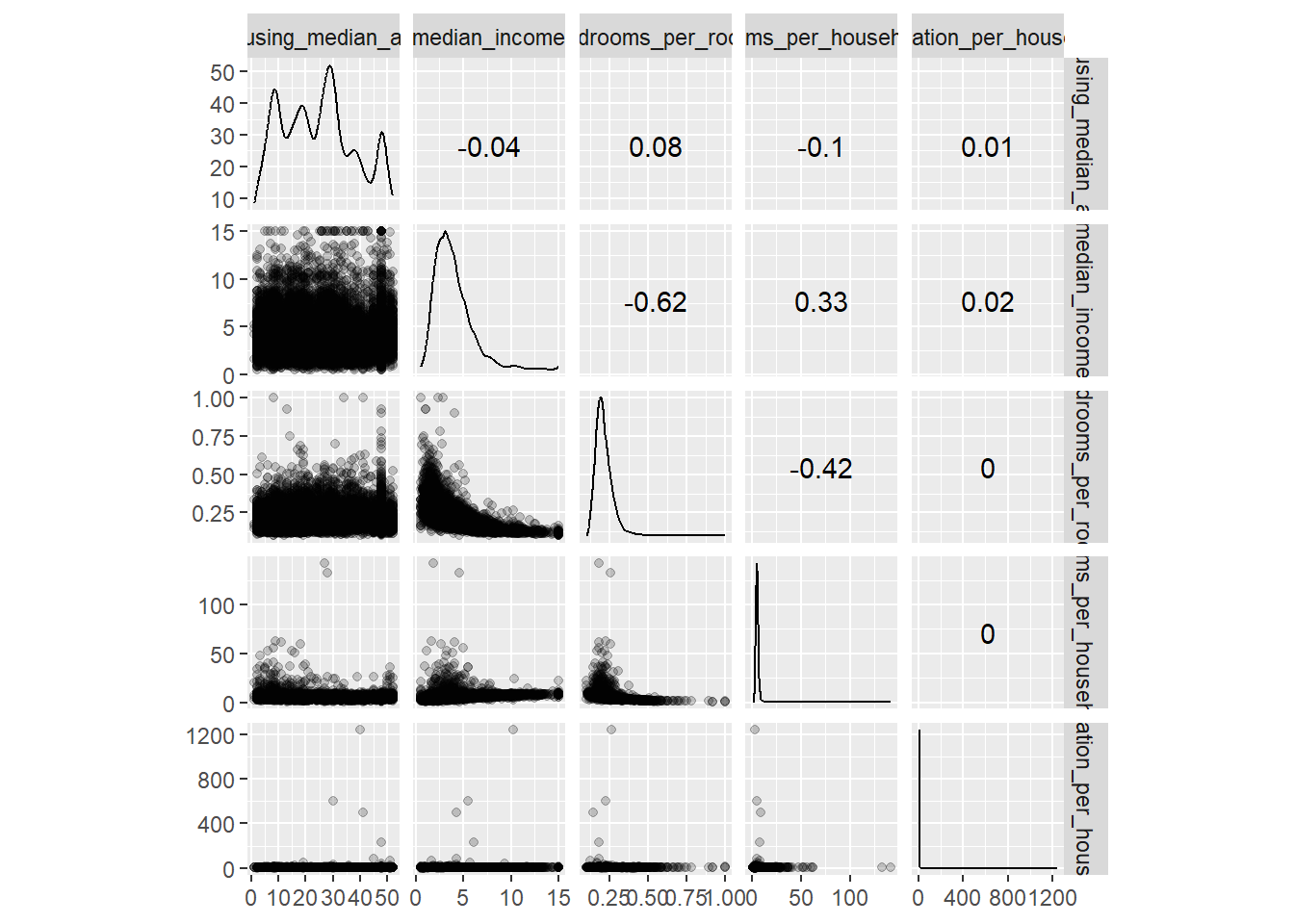
hdf |>
ggplot(aes(price_category)) + geom_bar()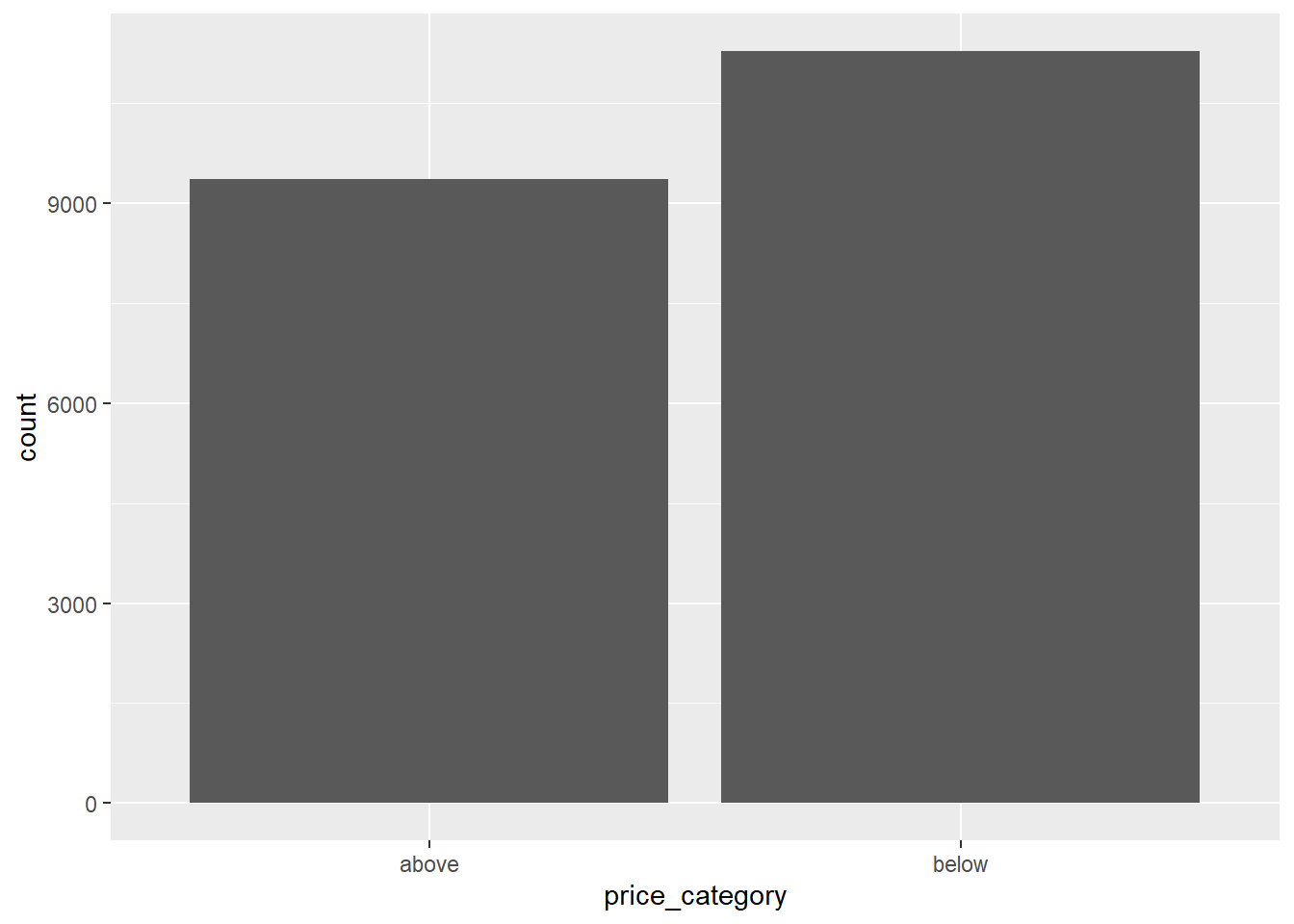
8.9 Your turn
Obtain hex binning scatterplot
Obtain scatterplot matrix using ggpairs
8.10 Split data into training and test
hdf_no_na <- hdf |>
drop_na()
set.seed(123)
data.split <- initial_split(hdf_no_na,
prop=3/4,
strata = price_category)
train.data <- training(data.split)
test.data <- testing(data.split)8.11 Build a randomforest model
library(randomForest)randomForest 4.7-1.2Type rfNews() to see new features/changes/bug fixes.
Attaching package: 'randomForest'The following object is masked from 'package:ggplot2':
marginThe following object is masked from 'package:dplyr':
combinerf <- randomForest(price_category ~ .,
data=train.data,
importance=TRUE)
test.data$predict <- predict(rf, test.data)
View(test.data)8.12 Accuracy measures
library(caret)Loading required package: lattice
Attaching package: 'caret'The following objects are masked from 'package:yardstick':
precision, recall, sensitivity, specificityThe following object is masked from 'package:purrr':
liftconfusionMatrix(data = test.data$predict,
reference = test.data$price_category)Confusion Matrix and Statistics
Reference
Prediction above below
above 1845 319
below 470 2475
Accuracy : 0.8456
95% CI : (0.8354, 0.8554)
No Information Rate : 0.5469
P-Value [Acc > NIR] : < 2.2e-16
Kappa : 0.6866
Mcnemar's Test P-Value : 9.287e-08
Sensitivity : 0.7970
Specificity : 0.8858
Pos Pred Value : 0.8526
Neg Pred Value : 0.8404
Prevalence : 0.4531
Detection Rate : 0.3611
Detection Prevalence : 0.4236
Balanced Accuracy : 0.8414
'Positive' Class : above
Sensitivity:…………………
Specificity:…………………
Prevalence:………………….
Positive Predictive Value:…………………
Negative Predictive Value:…………………
Detection rate:…………….
Detection prevalence:………………..
Balanced accuracy:……………………
Precision:……………….
Recall:…………………
F1 score:………………….
8.13 Exercise
Prepare a model to predict medv - Median value of owner-occupied homes in $1000s
R code to load data:
library(mlbench)
data(BostonHousing)
head(BostonHousing) crim zn indus chas nox rm age dis rad tax ptratio b lstat
1 0.00632 18 2.31 0 0.538 6.575 65.2 4.0900 1 296 15.3 396.90 4.98
2 0.02731 0 7.07 0 0.469 6.421 78.9 4.9671 2 242 17.8 396.90 9.14
3 0.02729 0 7.07 0 0.469 7.185 61.1 4.9671 2 242 17.8 392.83 4.03
4 0.03237 0 2.18 0 0.458 6.998 45.8 6.0622 3 222 18.7 394.63 2.94
5 0.06905 0 2.18 0 0.458 7.147 54.2 6.0622 3 222 18.7 396.90 5.33
6 0.02985 0 2.18 0 0.458 6.430 58.7 6.0622 3 222 18.7 394.12 5.21
medv
1 24.0
2 21.6
3 34.7
4 33.4
5 36.2
6 28.78.14 Data Profiling
skimr::skim(BostonHousing)| Name | BostonHousing |
| Number of rows | 506 |
| Number of columns | 14 |
| _______________________ | |
| Column type frequency: | |
| factor | 1 |
| numeric | 13 |
| ________________________ | |
| Group variables | None |
Variable type: factor
| skim_variable | n_missing | complete_rate | ordered | n_unique | top_counts |
|---|---|---|---|---|---|
| chas | 0 | 1 | FALSE | 2 | 0: 471, 1: 35 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| crim | 0 | 1 | 3.61 | 8.60 | 0.01 | 0.08 | 0.26 | 3.68 | 88.98 | ▇▁▁▁▁ |
| zn | 0 | 1 | 11.36 | 23.32 | 0.00 | 0.00 | 0.00 | 12.50 | 100.00 | ▇▁▁▁▁ |
| indus | 0 | 1 | 11.14 | 6.86 | 0.46 | 5.19 | 9.69 | 18.10 | 27.74 | ▇▆▁▇▁ |
| nox | 0 | 1 | 0.55 | 0.12 | 0.38 | 0.45 | 0.54 | 0.62 | 0.87 | ▇▇▆▅▁ |
| rm | 0 | 1 | 6.28 | 0.70 | 3.56 | 5.89 | 6.21 | 6.62 | 8.78 | ▁▂▇▂▁ |
| age | 0 | 1 | 68.57 | 28.15 | 2.90 | 45.02 | 77.50 | 94.07 | 100.00 | ▂▂▂▃▇ |
| dis | 0 | 1 | 3.80 | 2.11 | 1.13 | 2.10 | 3.21 | 5.19 | 12.13 | ▇▅▂▁▁ |
| rad | 0 | 1 | 9.55 | 8.71 | 1.00 | 4.00 | 5.00 | 24.00 | 24.00 | ▇▂▁▁▃ |
| tax | 0 | 1 | 408.24 | 168.54 | 187.00 | 279.00 | 330.00 | 666.00 | 711.00 | ▇▇▃▁▇ |
| ptratio | 0 | 1 | 18.46 | 2.16 | 12.60 | 17.40 | 19.05 | 20.20 | 22.00 | ▁▃▅▅▇ |
| b | 0 | 1 | 356.67 | 91.29 | 0.32 | 375.38 | 391.44 | 396.22 | 396.90 | ▁▁▁▁▇ |
| lstat | 0 | 1 | 12.65 | 7.14 | 1.73 | 6.95 | 11.36 | 16.96 | 37.97 | ▇▇▅▂▁ |
| medv | 0 | 1 | 22.53 | 9.20 | 5.00 | 17.02 | 21.20 | 25.00 | 50.00 | ▂▇▅▁▁ |